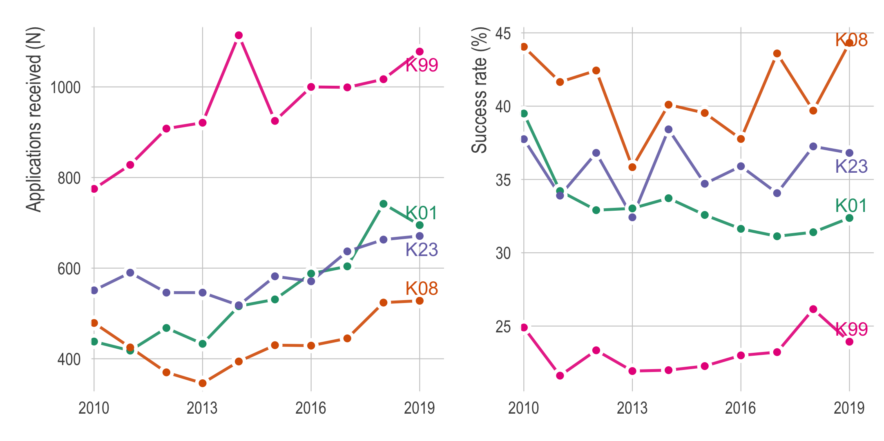
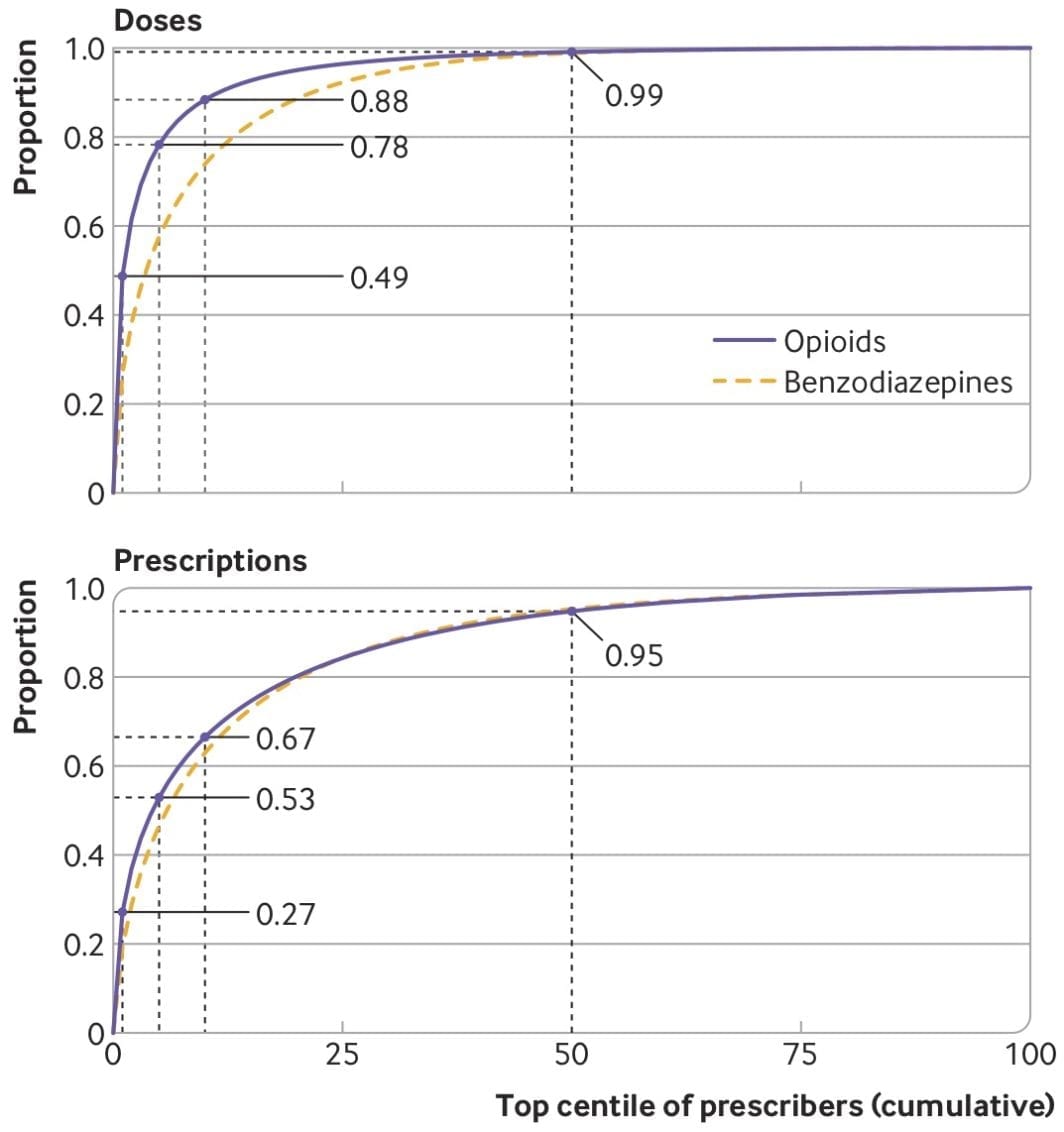
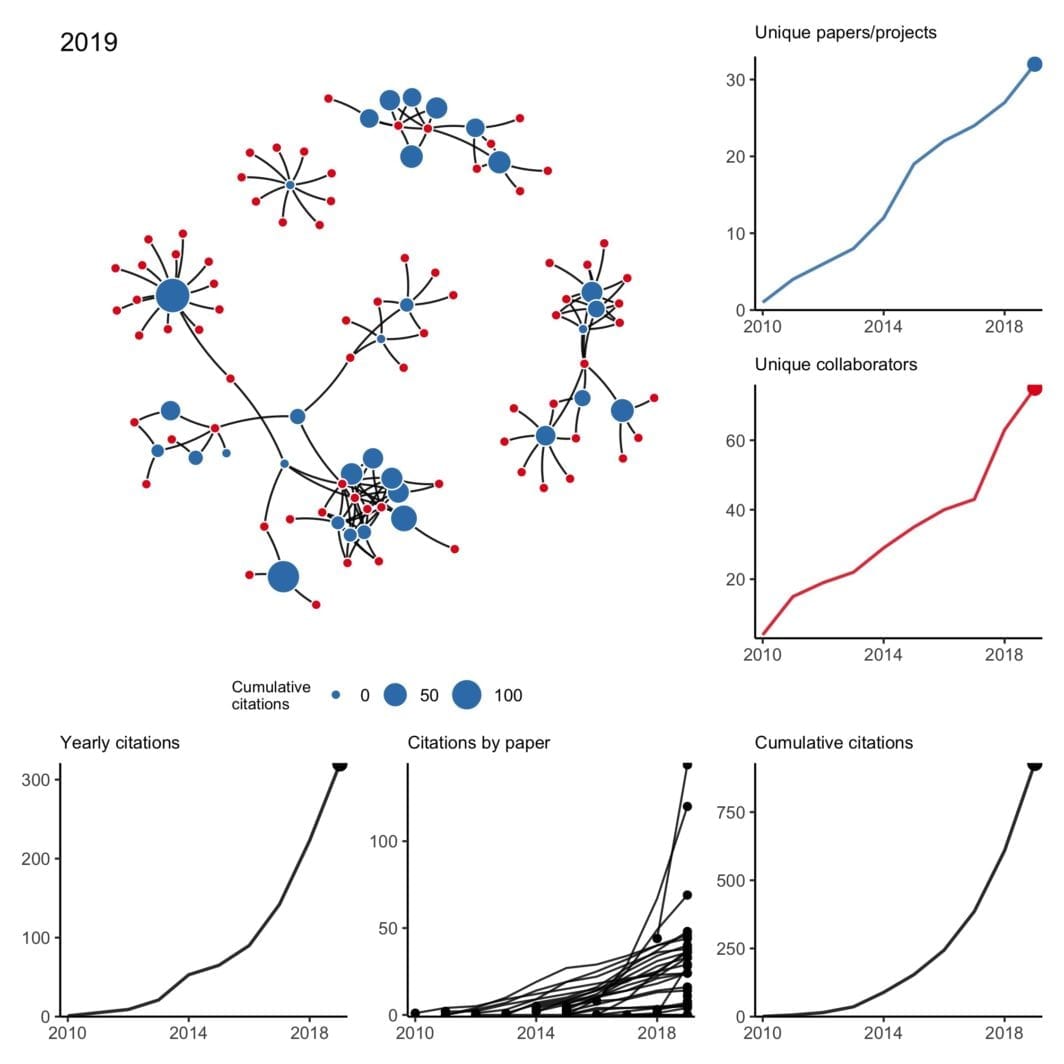
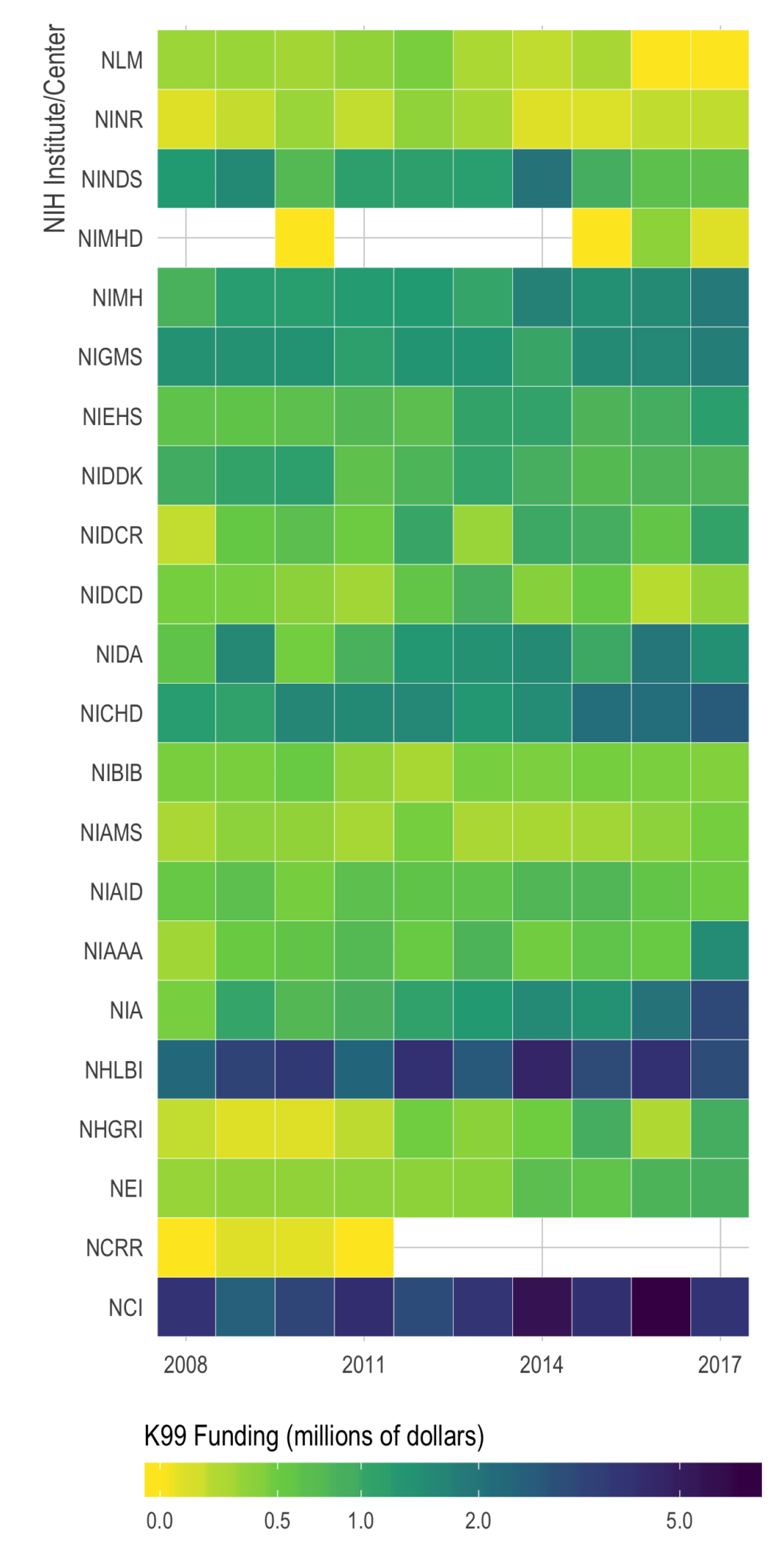
Here, I will try to pay it forward by sharing some thoughts and advice. There are plenty of good blog posts about applying for K99’s, so I’ll try to avoid repeating those. Instead, I’m going to focus on things I didn’t know before and/or didn’t read elsewhere. It will be based on (1) insight from others who applied, (2) advice from mentors of successfully funded applicants, and (3) my interpretation from reading about 20 K-award summary statements and applications (both funded and unfunded).
If there’s enough interest in the topic, I might write about the writing process itself, but here I’m going to focus on things to do before you apply. The tl;dr is (1) consider non-K99 options, (2) apply early in your postdoc, (3) give yourself more time than you think you’ll need, (4) be strategic about your target institution, and (5) avoid easy critiques.